Workshop
Installation
# using Anaconda environments
conda install cooler matplotlib
# using pip
pip install cooler matplotlib
Data
| File | Description |
|---|---|
22Rv1.hg19.40kbp.txt.gz |
Raw contact counts for chr6 and chr7 of GSE118629 |
hg19.40kbp.bed |
hg19 genome binned into 40kbp bins |
The contact matrix data is stored in a sparse matrix coordinate (COO) format where $M_{i,j} = v$ for each row whose columns are $(i, j, v)$.
Steps
Convert raw counts to cooler format
cooler load --assembly hg19 -f coo --one-based hg19.40kbp.bed 22Rv1.hg19.40kbp.txt.gz 22Rv1.cool
Balance using ICE
cooler balance 22Rv1.cool
Plot
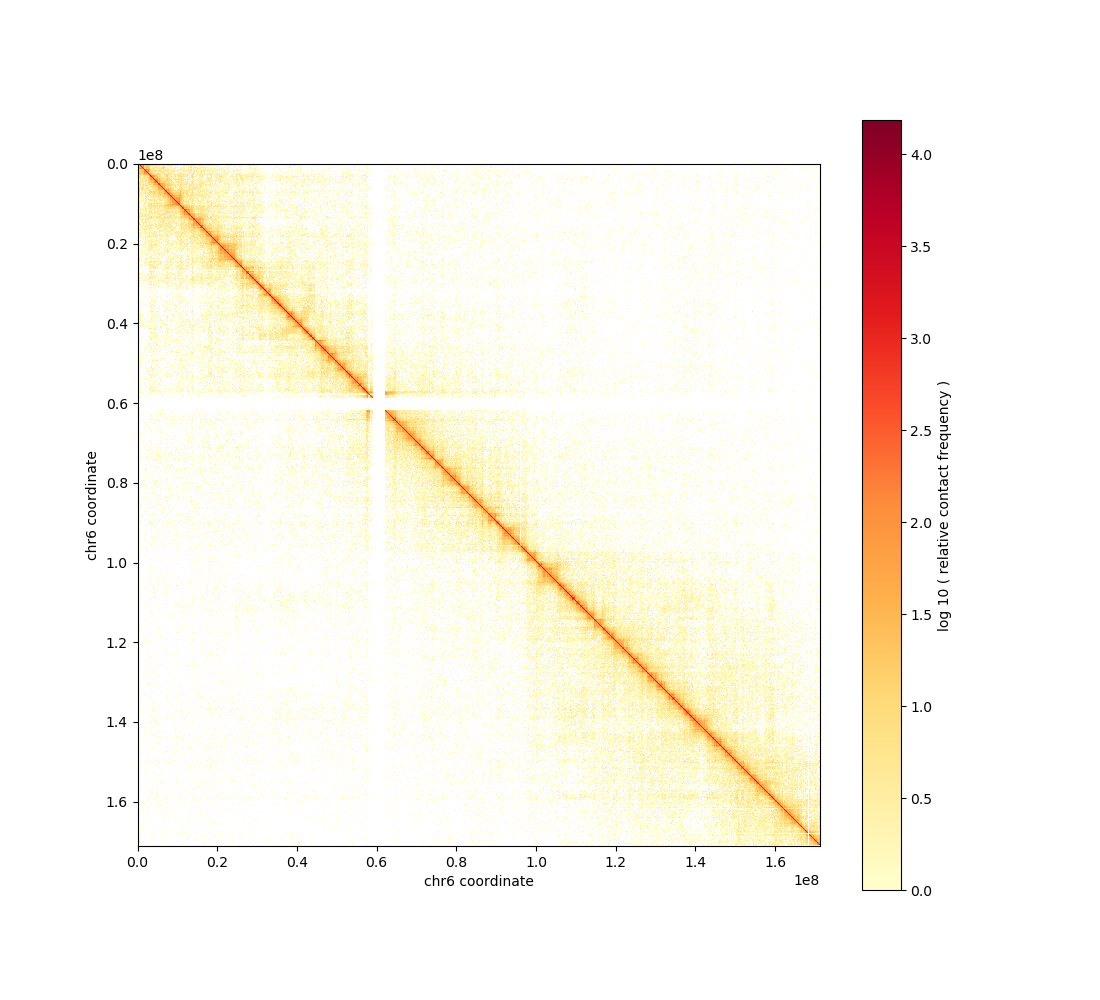
cooler show -o chr6.raw.png 22Rv1.cool chr6
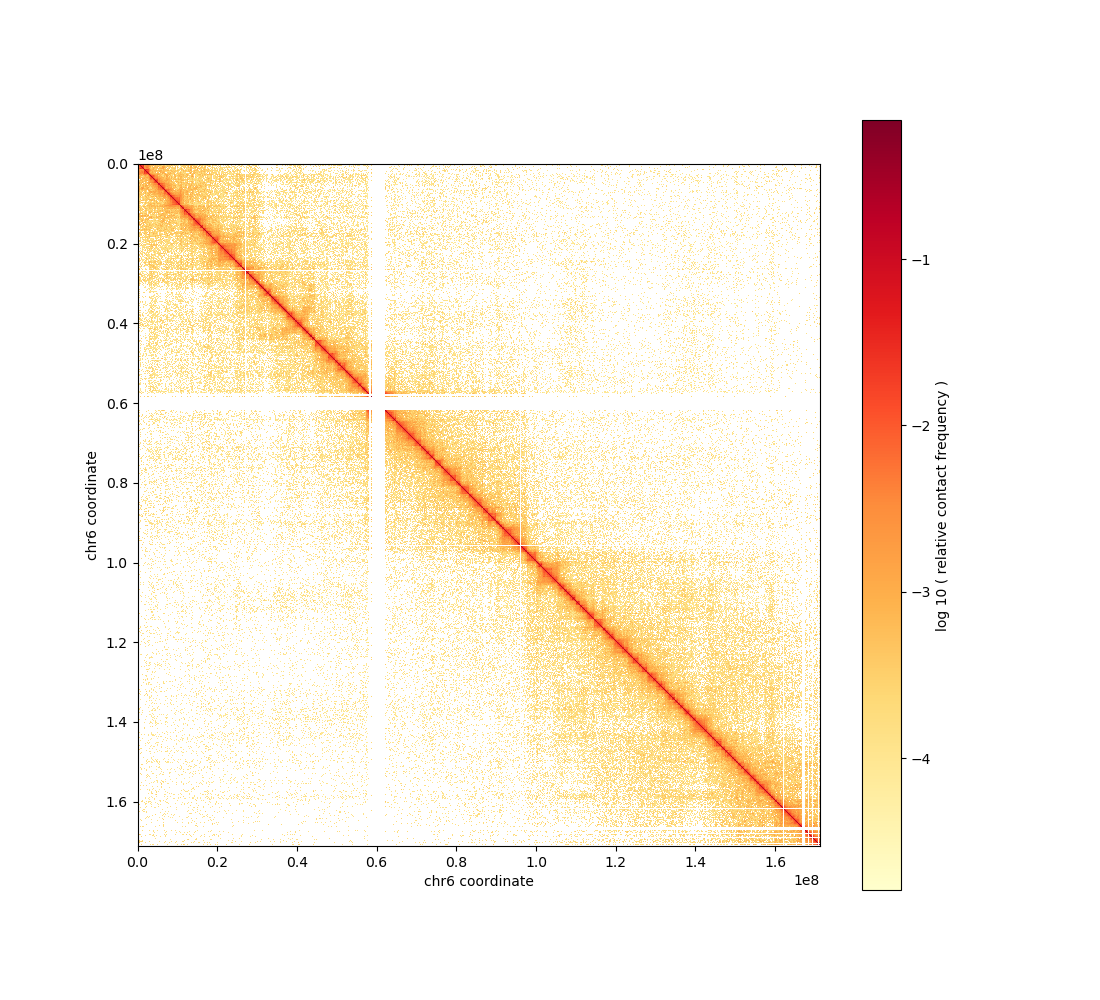
cooler show -b -o chr6.balanced.png 22Rv1.cool chr6
Raw chr6 contact matrix
Balanced chr6 contact matrix
Interactive exploration with Higlass
Higlass is a tool to interactively view HiC and other genomic data in a web browser.
You can see and interact with some example Higlass data here.